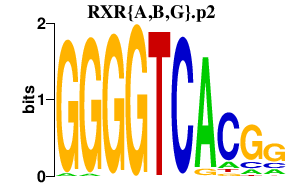
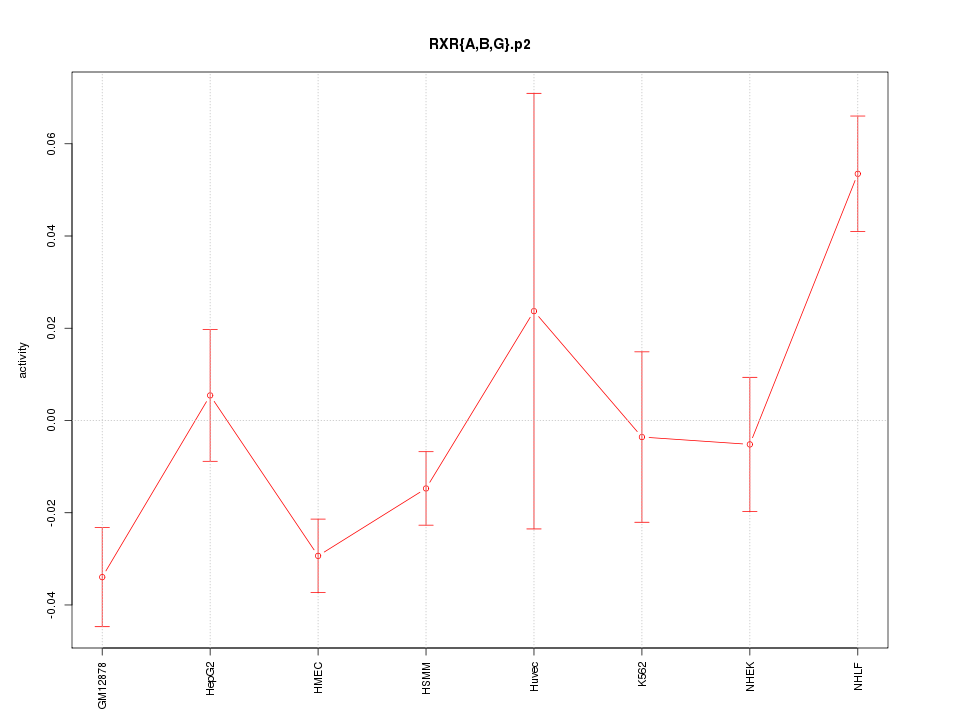
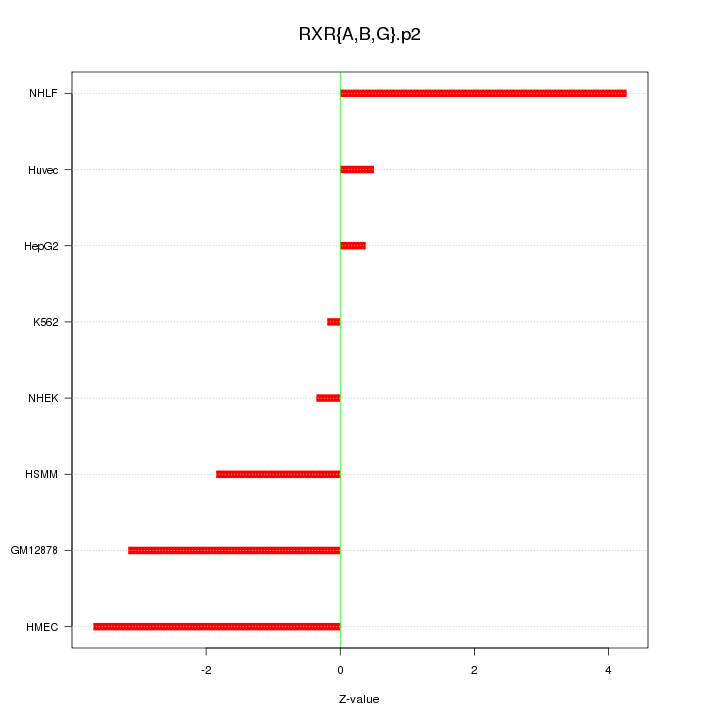
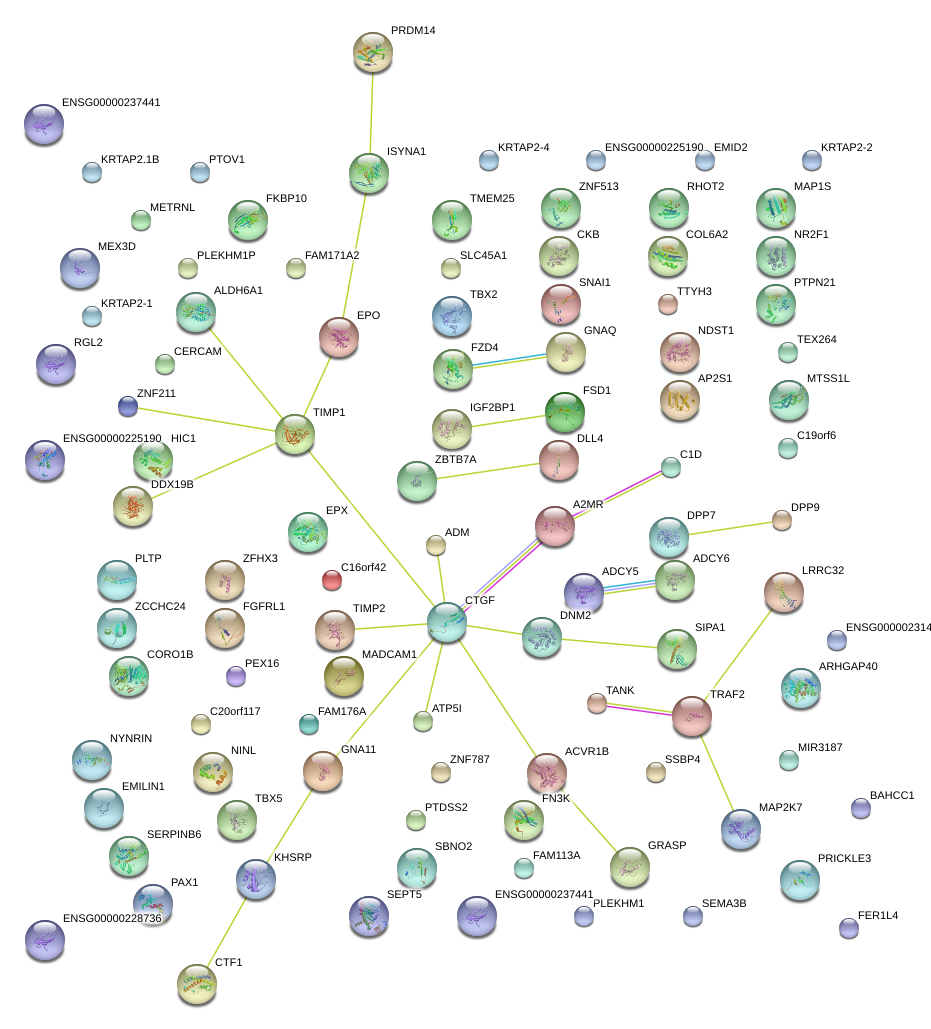
|
chr17_+_44429748
|
2.557
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr19_-_1125257
|
2.421
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr7_+_2638108
|
2.403
|
NM_025250
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr22_+_18081968
|
2.160
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr7_+_100156358
|
2.106
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr17_+_76988134
|
2.070
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr12_+_50686990
|
2.070
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr19_+_55046229
|
1.980
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_+_55046088
|
1.977
|
NM_017432
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_-_1125201
|
1.963
|
|
|
|
|
chr7_+_2638156
|
1.954
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr17_+_78630793
|
1.949
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr16_-_69277355
|
1.844
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr17_+_78286687
|
1.719
|
NM_022158
|
FN3K
|
fructosamine 3 kinase
|
|
chr19_-_1125263
|
1.719
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr19_+_18391211
|
1.715
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr17_+_56832255
|
1.651
|
|
TBX2
|
T-box 2
|
|
chr6_-_2916786
|
1.648
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr17_+_56831951
|
1.595
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr17_-_74433048
|
1.510
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_-_6375536
|
1.482
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr6_-_2916280
|
1.469
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr19_-_772913
|
1.432
|
NM_024888
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr5_+_92946348
|
1.419
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr9_-_101621964
|
1.401
|
|
|
|
|
chr14_-_88090641
|
1.386
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr19_+_18391423
|
1.371
|
|
|
|
|
chr19_-_61324456
|
1.368
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr17_-_36475652
|
1.354
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr2_+_27155197
|
1.325
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr11_+_440260
|
1.293
|
NM_030783
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr11_+_65162114
|
1.290
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr20_+_36663865
|
1.284
|
NM_001164431
|
ARHGAP40
|
Rho GTPase activating protein 40
|
|
chr15_+_39008822
|
1.281
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr4_+_995609
|
1.265
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr10_-_80875097
|
1.259
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr20_+_48032918
|
1.222
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr3_+_51680261
|
1.220
|
NM_001129884
NM_015926
|
TEX264
|
testis expressed 264
|
|
chr17_-_40923907
|
1.220
|
NM_014798
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr6_-_132314004
|
1.208
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr4_+_995395
|
1.202
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr19_-_772966
|
1.192
|
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chrX_-_48929685
|
1.176
|
NM_006150
|
PRICKLE3
|
prickle homolog 3 (Drosophila)
|
|
chr16_+_658158
|
1.166
|
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr12_+_55808514
|
1.160
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr11_+_10283171
|
1.151
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr17_-_39796649
|
1.146
|
NM_198475
|
FAM171A2
|
family with sequence similarity 171, member A2
|
|
chr6_-_33374275
|
1.145
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr20_-_25514137
|
1.138
|
NM_025176
|
NINL
|
ninein-like
|
|
chr16_+_30815428
|
1.132
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr9_+_130224950
|
1.119
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr14_-_103058568
|
1.114
|
|
CKB
|
creatine kinase, brain
|
|
chr17_+_37222731
|
1.106
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr4_-_658035
|
1.101
|
NM_007100
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E
|
|
chr11_+_440346
|
1.092
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr11_-_86343737
|
1.088
|
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr19_+_17691302
|
1.075
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr5_+_149845503
|
1.073
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr2_-_75641587
|
1.071
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr19_-_6375804
|
1.057
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr11_+_117906965
|
1.035
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr9_-_139128979
|
1.028
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr12_-_50502474
|
1.026
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|
|
chr19_-_6375782
|
1.024
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr11_-_76059435
|
1.015
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr3_+_50281582
|
1.014
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr19_+_7874774
|
1.011
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr20_-_2769245
|
1.010
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr19_+_447453
|
0.996
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr21_+_45649719
|
0.995
|
|
|
|
|
chr2_-_75641535
|
0.991
|
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr2_-_27457088
|
0.981
|
NM_144631
|
ZNF513
|
zinc finger protein 513
|
|
chr1_+_8306976
|
0.976
|
NM_001080397
|
SLC45A1
|
solute carrier family 45, member 1
|
|
chr20_-_2769120
|
0.975
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr17_-_36456949
|
0.970
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr17_+_1905104
|
0.965
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr19_-_18409971
|
0.965
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr16_-_71650034
|
0.960
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr12_-_113328271
|
0.957
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr19_+_7874765
|
0.957
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr12_+_50631734
|
0.956
|
|
ACVR1B
|
activin A receptor, type IB
|
|
chr2_+_27154938
|
0.954
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr19_-_18409788
|
0.954
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr11_-_86343859
|
0.953
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr19_-_4016467
|
0.936
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr19_-_972122
|
0.936
|
NM_001033026
NM_033420
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr20_+_21634515
|
0.930
|
|
PAX1
|
paired box 1
|
|
chr8_-_71146022
|
0.924
|
|
PRDM14
|
PR domain containing 14
|
|
chr19_-_1518875
|
0.919
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr11_-_66967481
|
0.910
|
|
CORO1B
|
coronin, actin binding protein, 1B
|
|
chr8_-_71146103
|
0.910
|
NM_024504
|
PRDM14
|
PR domain containing 14
|
|
chr19_-_971995
|
0.899
|
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr11_-_45896144
|
0.896
|
NM_004813
NM_057174
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr17_-_40923864
|
0.894
|
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr20_-_43972949
|
0.892
|
|
PLTP
|
phospholipid transfer protein
|
|
chr16_-_1341766
|
0.878
|
NM_001001410
|
C16orf42
|
chromosome 16 open reading frame 42
|
|
chr19_+_7874762
|
0.876
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr19_+_7874764
|
0.875
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr19_+_7874820
|
0.869
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr21_+_46342440
|
0.864
|
NM_001849
NM_058174
NM_058175
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr20_-_33658823
|
0.863
|
|
FER1L4
|
fer-1-like 4 (C. elegans) pseudogene
|
|
chr14_+_23937766
|
0.862
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr7_+_100792841
|
0.856
|
NM_133457
|
EMID2
|
EMI domain containing 2
|
|
chr20_-_34925483
|
0.856
|
NM_080627
|
KIAA0889
|
KIAA0889
|
|
chr19_+_4255590
|
0.848
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr19_-_4674814
|
0.848
|
NM_139159
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr19_+_55045754
|
0.838
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr22_+_18085891
|
0.833
|
|
SEPT5
|
septin 5
|
|
chr20_+_21634293
|
0.832
|
NM_006192
|
PAX1
|
paired box 1
|
|
chr19_+_3045528
|
0.827
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr3_-_124650081
|
0.826
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr19_-_52045890
|
0.822
|
NM_004069
NM_021575
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr19_-_6375820
|
0.822
|
NM_003685
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr9_+_138900760
|
0.822
|
NM_021138
|
TRAF2
|
TNF receptor-associated factor 2
|
|
chr17_+_37222487
|
0.818
|
NM_021939
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr1_-_36562341
|
0.811
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr14_+_23654161
|
0.808
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr19_+_17047569
|
0.805
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr17_-_75808680
|
0.803
|
NM_000199
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr21_-_32879617
|
0.801
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr9_+_35480018
|
0.795
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr3_-_49178762
|
0.795
|
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr5_-_131591380
|
0.794
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr8_-_93184629
|
0.793
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_37222726
|
0.792
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr17_+_7151653
|
0.791
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr16_-_73857214
|
0.791
|
NM_001170720
NM_001170714
NM_001170718
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr3_-_49178773
|
0.788
|
NM_022903
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr17_+_39620099
|
0.787
|
NM_177441
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr22_+_18085990
|
0.786
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chrX_-_30237403
|
0.783
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr19_+_38376980
|
0.779
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr17_+_35472566
|
0.776
|
NM_001190918
NM_003250
NM_199334
|
THRA
|
thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian)
|
|
chr7_+_78920846
|
0.774
|
|
LOC100505881
|
hypothetical LOC100505881
|
|
chr19_+_59064471
|
0.773
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr10_-_103444899
|
0.772
|
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr19_-_5291700
|
0.772
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr11_+_440408
|
0.771
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr17_-_76985538
|
0.769
|
|
|
|
|
chr3_-_49178738
|
0.768
|
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr20_-_33489383
|
0.766
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr19_-_4674791
|
0.766
|
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr17_+_39620108
|
0.764
|
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr9_+_136673534
|
0.763
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr9_+_35479989
|
0.763
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_+_39619826
|
0.762
|
NM_001076674
NM_024107
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr9_+_130222511
|
0.759
|
NM_016174
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr11_+_117907083
|
0.757
|
|
TMEM25
|
transmembrane protein 25
|
|
chr1_+_1233834
|
0.753
|
NM_153339
|
PUSL1
|
pseudouridylate synthase-like 1
|
|
chr17_+_35471971
|
0.752
|
NM_001190919
|
THRA
|
thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian)
|
|
chr19_-_63761559
|
0.748
|
|
UBE2M
|
ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast)
|
|
chrX_-_48929789
|
0.745
|
|
PRICKLE3
|
prickle homolog 3 (Drosophila)
|
|
chr6_-_33374678
|
0.743
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr17_-_28644118
|
0.741
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr11_+_66381451
|
0.739
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr19_-_844184
|
0.738
|
|
MED16
|
mediator complex subunit 16
|
|
chr19_+_17691283
|
0.734
|
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr9_+_115970599
|
0.733
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr16_+_66129481
|
0.732
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr19_-_6230927
|
0.726
|
NM_005934
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr19_+_2279679
|
0.710
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr20_+_2769338
|
0.709
|
NM_022575
NM_080413
|
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae)
|
|
chr19_-_14089560
|
0.708
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr1_+_9571518
|
0.707
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr4_+_134292630
|
0.705
|
|
PCDH10
|
protocadherin 10
|
|
chr19_+_7874701
|
0.702
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr9_-_138454071
|
0.701
|
NM_019892
|
INPP5E
|
inositol polyphosphate-5-phosphatase, 72 kDa
|
|
chr16_+_56327127
|
0.700
|
NM_005886
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr16_+_658081
|
0.699
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr20_-_61933012
|
0.699
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr11_+_66381077
|
0.697
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr9_-_16860664
|
0.694
|
|
BNC2
|
basonuclin 2
|
|
chr19_+_3457264
|
0.693
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr9_-_130573671
|
0.689
|
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr19_+_47480445
|
0.689
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr19_+_45664889
|
0.688
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr9_+_115678365
|
0.686
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr1_-_153213483
|
0.686
|
NM_001130041
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr22_+_18485095
|
0.682
|
|
RANBP1
|
RAN binding protein 1
|
|
chr16_-_11257539
|
0.680
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr1_-_32460526
|
0.678
|
|
TMEM234
|
transmembrane protein 234
|
|
chr20_-_43972982
|
0.678
|
|
PLTP
|
phospholipid transfer protein
|
|
chr9_+_130942029
|
0.676
|
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr17_-_7061641
|
0.676
|
NM_001128827
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr17_-_23716391
|
0.676
|
NM_001080837
|
SEBOX
|
SEBOX homeobox
|
|
chr14_+_76298693
|
0.672
|
|
VASH1
|
vasohibin 1
|
|
chr5_-_149809424
|
0.671
|
|
RPS14
|
ribosomal protein S14
|
|
chr17_-_74432671
|
0.670
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr16_+_56327168
|
0.670
|
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr11_+_47227009
|
0.668
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr19_+_59064795
|
0.668
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr9_-_139128777
|
0.666
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr17_-_37221976
|
0.662
|
NM_006455
|
LEPREL4
|
leprecan-like 4
|
|
chr14_+_22375632
|
0.662
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr16_+_68705714
|
0.661
|
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr9_-_109291575
|
0.660
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr14_-_60260211
|
0.659
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr19_+_54723044
|
0.655
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr7_-_75461897
|
0.653
|
NM_031925
|
TMEM120A
|
transmembrane protein 120A
|
|
chr3_-_48647811
|
0.653
|
|
SLC26A6
|
solute carrier family 26, member 6
|